This article describes the essentials of R coding style best practices. It’s based on the tidyverse style guide. Google’s current guide is also derived from the tidyverse style guide.
Two importants R packages are available to help you in applying the R coding style best practices:
- styler allows you to interactively restyle selected text, files, or entire projects. It includes an RStudio add-in, the easiest way to re-style existing code.
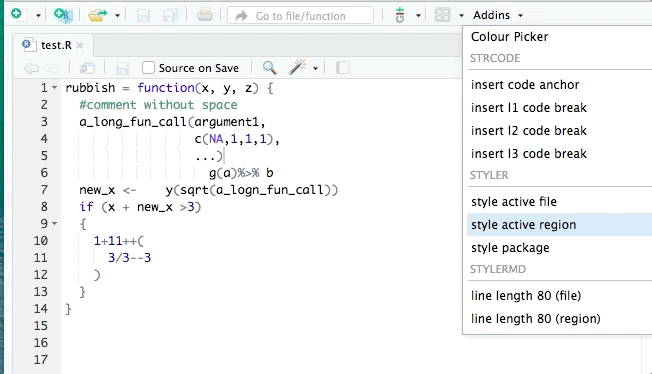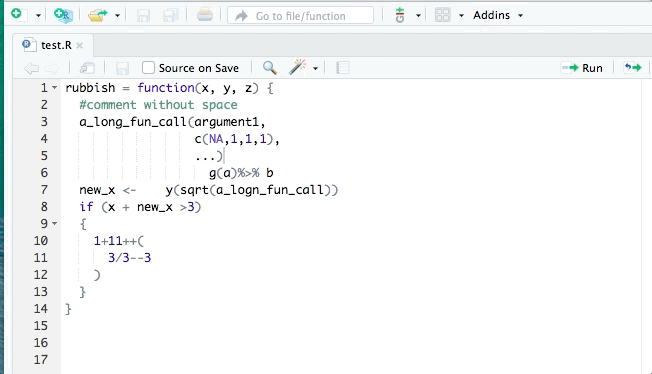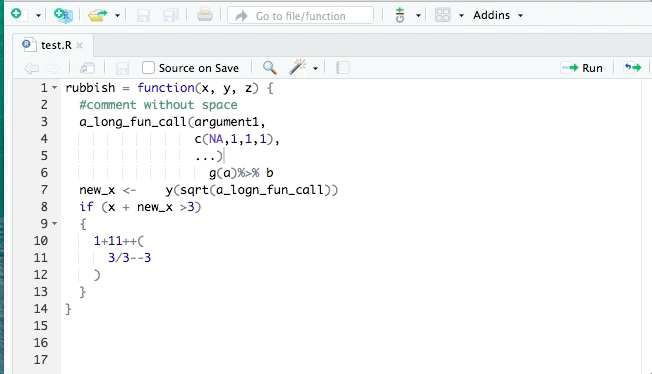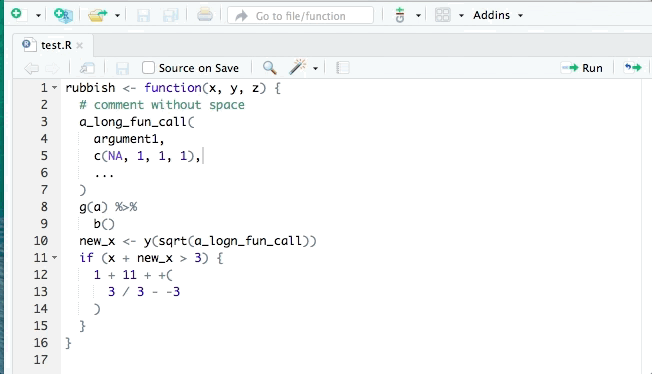
The goal of styler is to provide non-invasive pretty-printing of R source code while adhering to the tidyverse formatting rules. It can be installed using the following R code: install.packages("styler"). Key functions are:
style_file(): styles .R, .Rmd .Rnw and .Rprofile, files.style_dir(): styles all .R and/or .Rmd files in a directory.style_pkg(): styles the source files of an R package.
The styler functionality is made available through other tools, most notably: usethis::use_tidy_style() styles your project according to the tidyverse style guide.
- lintr performs automated checks to confirm that you conform to the style guide. It can be installed using the following R code:
install.packages("lintr").

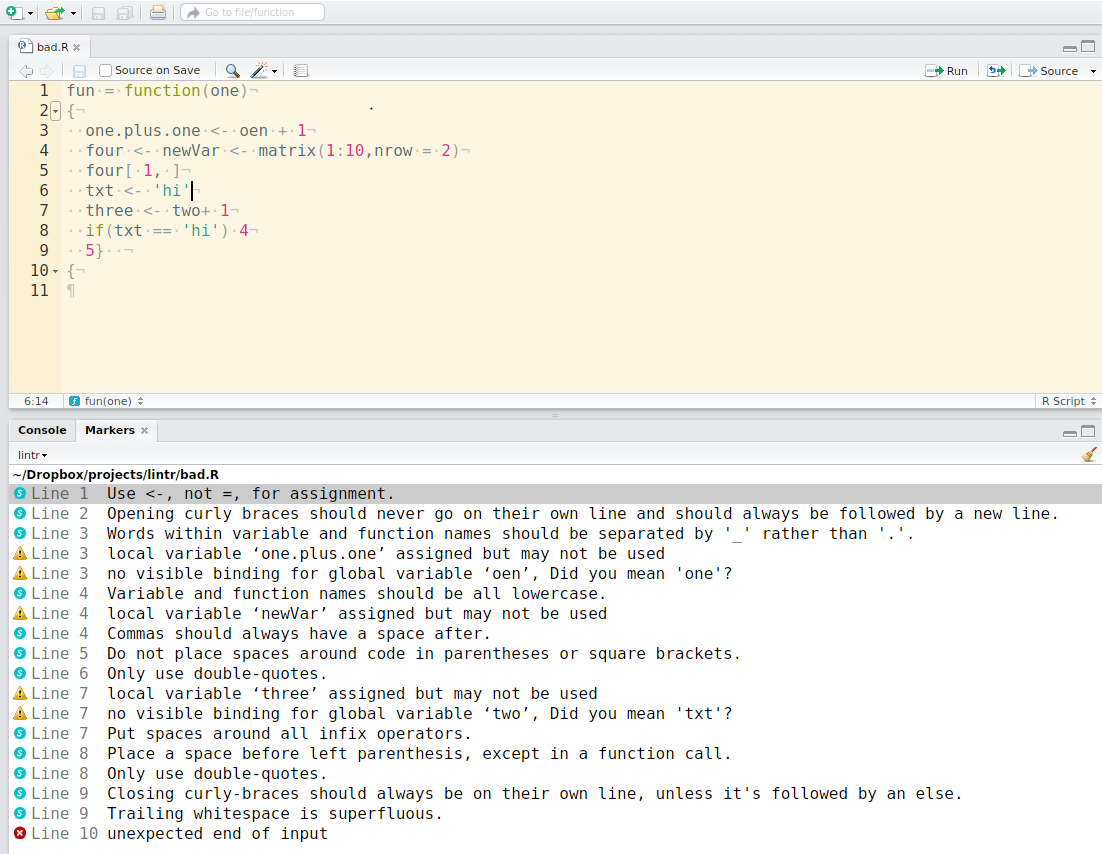
The lintr lints are automatically displayed in the RStudio Markers pane (Rstudio versions > v0.99.206). In order to show the “Markers” pane in RStudio: Menu “Tools” -> “Global Options…”, a window with title “Options” will pop up. In that window: Click “Code” on the left; Click “Diagnostics” tab; check “Show diagnostics for R”.
To lint a source file test.R type in the Console lintr::lint("test.R") and look at the result in the “Markers” pane.
This package also includes two addins for linting the current source and package. To bind the addin to a keyboard shortcut navigate to Tools > addins > Browse Addins > Keyboard Shortcuts. It’s recommended to use Alt+Shift+L for linting the current source code and Ctrl+Shift+Alt+L to code the package. These are easy to remember as you are Alt+Shift+L(int) 😉
In this tutorial, you will learn best practices for:
- File naming and content structures
- Variables and object naming conventions
- R syntax and pipes best practices
Contents:
Files
File naming conventions
File names should be meaningful and end in .R. Avoid using special characters in file names - stick with numbers, letters, -, and _.
# Good
fit_models.R
utility_functions.R
# Bad
fit models.R
foo.r
stuff.rIf files should be run in a particular order, prefix them with numbers. If it seems likely you’ll have more than 10 files, left pad with zero:
00_download.R
01_explore.R
...
09_model.R
10_visualize.RIf you later realise that you’ve missed some steps, it’s tempting to use 02a, 02b, etc. However, it’s generally better to bite the bullet and rename all files.
File content structure
- Load all required packages at the very beginning of the file
- Use commented lines of
-and=to break up your file into easily readable chunks.
# Load data ---------------------------
# Plot data ---------------------------Syntax
Object naming convention
- Use only lowercase letters and numbers.
- Use underscores (
_) (so called snake case) to separate words within a name. - Use names that are concise and meaningful (this is not easy!).
- Generally, variable names should be nouns and function names should be verbs.
- Where possible, avoid re-using names of common functions and variables. This will cause confusion for the readers of your code.
- If you find yourself attempting to cram data into variable names (e.g.
model_2018,model_2019,model_2020), consider using a list or data frame instead.
# Examples of variable names ------------
# Good
day_one
day_1
# Bad
DayOne
dayoneSpacing
Commas
Always put a space after a comma, never before, just like in regular English.
# Good
x[, 1]
# Bad
x[,1]
x[ ,1]
x[ , 1]Parentheses
Do not put spaces inside or outside parentheses for regular function calls.
# Good
mean(x, na.rm = TRUE)
# Bad
mean (x, na.rm = TRUE)
mean( x, na.rm = TRUE )Place a space before and after () when used with if, for, or while.
# Good
if (debug) {
show(x)
}
# Bad
if(debug){
show(x)
}Place a space after () used for function arguments:
# Good
function(x) {}
# Bad
function (x) {}
function(x){}Infix operators
Most infix operators (==, +, -, <-, etc.) should always be surrounded by spaces:
# Good
height <- (feet * 12) + inches
mean(x, na.rm = 10)
# Bad
height<-feet*12+inches
mean(x, na.rm=10)There are a few exceptions, to this rule: ::, :::, $, @, [, [[, ^, unary -, unary +, and :.
# Good
sqrt(x^2 + y^2)
df$z
x <- 1:10
# Bad
sqrt(x ^ 2 + y ^ 2)
df $ z
x <- 1 : 10Extra spaces
Adding extra spaces ok if it improves alignment of = or <-.
# Good
list(
total = a + b + c,
mean = (a + b + c) / n
)
# Also fine
list(
total = a + b + c,
mean = (a + b + c) / n
)Do not add extra spaces to places where space is not usually allowed.
Argument names
A function’s arguments typically fall into two broad categories: one supplies the data to compute on; the other controls the details of computation. When you call a function, you typically omit the names of data arguments, because they are used so commonly. If you override the default value of an argument, use the full name:
# Good
mean(1:10, na.rm = TRUE)
# Bad
mean(x = 1:10, , FALSE)
mean(, TRUE, x = c(1:10, NA))Avoid partial matching.
Code blocks
Curly braces, {}, define the most important hierarchy of R code. To make this hierarchy easy to see:
{should be the last character on the line. Related code (e.g., anifclause, a function declaration, a trailing comma, …) must be on the same line as the opening brace.- The contents should be indented by two spaces.
}should be the first character on the line.
# Good
if (y < 0 && debug) {
message("y is negative")
}
if (y == 0) {
if (x > 0) {
log(x)
} else {
message("x is negative or zero")
}
} else {
y^x
}
# Bad
if (y < 0 && debug) {
message("Y is negative")
}
if (y == 0)
{
if (x > 0) {
log(x)
} else {
message("x is negative or zero")
}
} else { y ^ x }Long lines
- Strive to limit your code to 80 characters per line.
- If a function call is too long to fit on a single line, use one line each for the function name, each argument, and the closing
). This makes the code easier to read and to change later. - You can place several arguments on the same line if they are closely related to each other
# Good
do_something_very_complicated(
something = "that",
requires = many,
arguments = "some of which may be long"
)
# Bad
do_something_very_complicated("that", requires, many, arguments,
"some of which may be long"
)Assignment
Use <-, not =, for assignment.
# Good
x <- 5
# Bad
x = 5Semicolons
Don’t put ; at the end of a line, and don’t use ; to put multiple commands on one line.
Quotes
Use ", not ', for quoting text. The only exception is when the text already contains double quotes and no single quotes.
# Good
"Text"
'Text with "quotes"'
'<a href="http://style.tidyverse.org">A link</a>'
# Bad
'Text'
'Text with "double" and \'single\' quotes'Functions
Function naming convention
Use verbs for function names:
# Good
add_row()
permute()
# Bad
row_adder()
permutation()Long line function definition
If a function definition runs over multiple lines, indent the second line to where the definition starts.
# Good
long_function_name <- function(a = "a long argument",
b = "another argument",
c = "another long argument") {
# As usual code is indented by two spaces.
}
# Bad
long_function_name <- function(a = "a long argument",
b = "another argument",
c = "another long argument") {
# Here it's hard to spot where the definition ends and the
# code begins
}return() function
- Only use
return()for early returns. Otherwise, rely on R to return the result of the last evaluated expression. - Return statements should always be on their own line.
# Good
find_abs <- function(x) {
if (x > 0) {
return(x)
}
x * -1
}
add_two <- function(x, y) {
x + y
}
# Bad
add_two <- function(x, y) {
return(x + y)
}If your function is called primarily for its side-effects (like printing, plotting, or saving to disk), it should return the first argument invisibly. This makes it possible to use the function as part of a pipe. print methods should usually do this, like this example from httr:
print.url <- function(x, ...) {
cat("Url: ", build_url(x), "\n", sep = "")
invisible(x)
}Pipes
Introduction
Use %>% to emphasise a sequence of actions.
Avoid using the pipe when:
- You need to manipulate more than one object at a time. Reserve pipes for a sequence of steps applied to one primary object.
- There are meaningful intermediate objects that could be given informative names.
Whitespace
%>% should always have a space before it, and should usually be followed by a new line. After the first step, each line should be indented by two spaces.
# Good
iris %>%
group_by(Species) %>%
summarize_if(is.numeric, mean) %>%
ungroup() %>%
gather(measure, value, -Species) %>%
arrange(value)
# Bad
iris %>% group_by(Species) %>% summarize_all(mean) %>%
ungroup %>% gather(measure, value, -Species) %>%
arrange(value)Long lines
If the arguments to a function don’t all fit on one line, put each argument on its own line and indent:
iris %>%
group_by(Species) %>%
summarise(
Sepal.Length = mean(Sepal.Length),
Sepal.Width = mean(Sepal.Width),
Species = n_distinct(Species)
)Short pipes
A one-step pipe can stay on one line, but unless you plan to expand it later on, you should consider rewriting it to a regular function call.
# Good
iris %>% arrange(Species)
iris %>%
arrange(Species)
arrange(iris, Species)Recommended for you
This section contains best data science and self-development resources to help you on your path.
Books - Data Science
Our Books
- Practical Guide to Cluster Analysis in R by A. Kassambara (Datanovia)
- Practical Guide To Principal Component Methods in R by A. Kassambara (Datanovia)
- Machine Learning Essentials: Practical Guide in R by A. Kassambara (Datanovia)
- R Graphics Essentials for Great Data Visualization by A. Kassambara (Datanovia)
- GGPlot2 Essentials for Great Data Visualization in R by A. Kassambara (Datanovia)
- Network Analysis and Visualization in R by A. Kassambara (Datanovia)
- Practical Statistics in R for Comparing Groups: Numerical Variables by A. Kassambara (Datanovia)
- Inter-Rater Reliability Essentials: Practical Guide in R by A. Kassambara (Datanovia)
Others
- R for Data Science: Import, Tidy, Transform, Visualize, and Model Data by Hadley Wickham & Garrett Grolemund
- Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems by Aurelien Géron
- Practical Statistics for Data Scientists: 50 Essential Concepts by Peter Bruce & Andrew Bruce
- Hands-On Programming with R: Write Your Own Functions And Simulations by Garrett Grolemund & Hadley Wickham
- An Introduction to Statistical Learning: with Applications in R by Gareth James et al.
- Deep Learning with R by François Chollet & J.J. Allaire
- Deep Learning with Python by François Chollet
Version:
 Français
Français







Comments
If you need comments to explain what your code is doing, consider rewriting your code to be clearer. If you discover that you have more comments than code, consider switching to R Markdown.