This chapter describes how to transform data to normal distribution in R. Parametric methods, such as t-test and ANOVA tests, assume that the dependent (outcome) variable is approximately normally distributed for every groups to be compared.
In the situation where the normality assumption is not met, you could consider transform the data for correcting the non-normal distributions.
When dealing with t-test and ANOVA assumptions, you just need to transform the dependent variable. However, when dealing with the assumptions of linear regression, you can consider transformations of either the independent or dependent variable or both for achieving a linear relationship between variables or to make sure there is homoscedasticity.
Note that, transformation will not always be successful.
In this article, you will learn:
- Common types of non-normal distributions
- Methods for transforming the data to correct the non-normal distributions
Contents:
Related Book
Practical Statistics in R II - Comparing Groups: Numerical VariablesNon-normal distributions
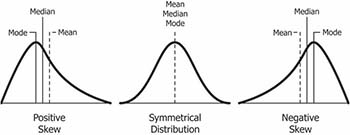
Skewness is a measure of symmetry for a distribution. The value can be positive, negative or undefined. In a skewed distribution, the central tendency measures (mean, median, mode) will not be equal.
(Image courtesy: https://www.safaribooksonline.com/library/view/clojure-for-data/9781784397180/ch01s13.html)
- Positively skewed distribution (or right skewed): The most frequent values are low; tail is toward the high values (on the right-hand side). Generally,
Mode < Median < Mean. - Negatively skewed distribution (or left skewed), the most frequent values are high; tail is toward low values (on the left-hand side). Generally,
Mode > Median > Mean.
The direction of skewness is given by the sign of the skewness coefficient:
- A zero means no skewness at all (normal distribution).
- A negative value means the distribution is negatively skewed.
- A positive value means the distribution is positively skewed.
The larger the value of skewness, the larger the distribution differs from a normal distribution.
The skewness coefficient can be computed using the moments R packages:
library(moments)
skewness(iris$Sepal.Length, na.rm = TRUE)## [1] 0.312Transformation methods
This section describes different transformation methods, depending to the type of normality violation.
Some common heuristics transformations for non-normal data include:
- square-root for moderate skew:
sqrt(x)for positively skewed data,sqrt(max(x+1) - x)for negatively skewed data
- log for greater skew:
log10(x)for positively skewed data,log10(max(x+1) - x)for negatively skewed data
- inverse for severe skew:
1/xfor positively skewed data1/(max(x+1) - x)for negatively skewed data
- Linearity and heteroscedasticity:
- first try
logtransformation in a situation where the dependent variable starts to increase more rapidly with increasing independent variable values - If your data does the opposite – dependent variable values decrease more rapidly with increasing independent variable values – you can first consider a
squaretransformation.
- first try
Note that, when using a log transformation, a constant should be added to all values to make them all positive before transformation.
Examples of transforming skewed data
Prerequisites
Make sure you have installed the following R packages:
ggpubrfor creating easily publication ready plotsmomentsfor computing skewness
Start by loading the packages:
library(ggpubr)
library(moments)Demo dataset: Built-in R dataset USJudgeRatings.
data("USJudgeRatings")
df <- USJudgeRatings
head(df)## CONT INTG DMNR DILG CFMG DECI PREP FAMI ORAL WRIT PHYS RTEN
## AARONSON,L.H. 5.7 7.9 7.7 7.3 7.1 7.4 7.1 7.1 7.1 7.0 8.3 7.8
## ALEXANDER,J.M. 6.8 8.9 8.8 8.5 7.8 8.1 8.0 8.0 7.8 7.9 8.5 8.7
## ARMENTANO,A.J. 7.2 8.1 7.8 7.8 7.5 7.6 7.5 7.5 7.3 7.4 7.9 7.8
## BERDON,R.I. 6.8 8.8 8.5 8.8 8.3 8.5 8.7 8.7 8.4 8.5 8.8 8.7
## BRACKEN,J.J. 7.3 6.4 4.3 6.5 6.0 6.2 5.7 5.7 5.1 5.3 5.5 4.8
## BURNS,E.B. 6.2 8.8 8.7 8.5 7.9 8.0 8.1 8.0 8.0 8.0 8.6 8.6In the following examples, we’ll consider two variables:
CONT: Number of contacts of lawyer with judge. Positively skewed.PHYS: Physical ability. Negatively skewed
Visualization
Plot the density distribution of each variable and compare the observed distribution to what we would expect if it were perfectly normal (dashed red line).
# Distribution of CONT variable
ggdensity(df, x = "CONT", fill = "lightgray", title = "CONT") +
scale_x_continuous(limits = c(3, 12)) +
stat_overlay_normal_density(color = "red", linetype = "dashed")
# Distribution of PHYS variable
ggdensity(df, x = "PHYS", fill = "lightgray", title = "PHYS") +
scale_x_continuous(limits = c(3, 12)) +
stat_overlay_normal_density(color = "red", linetype = "dashed")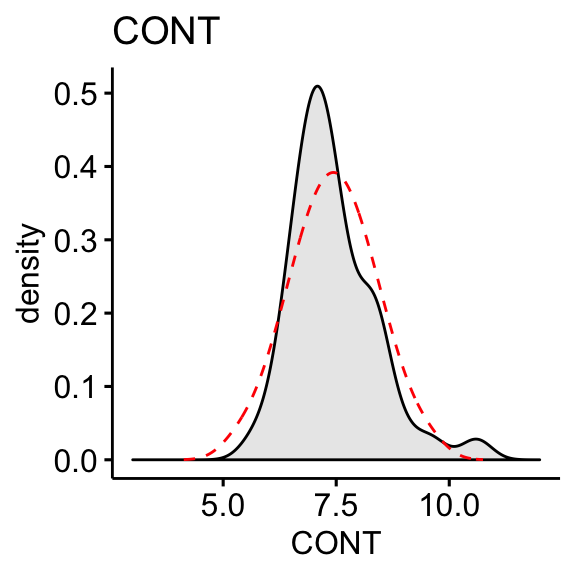
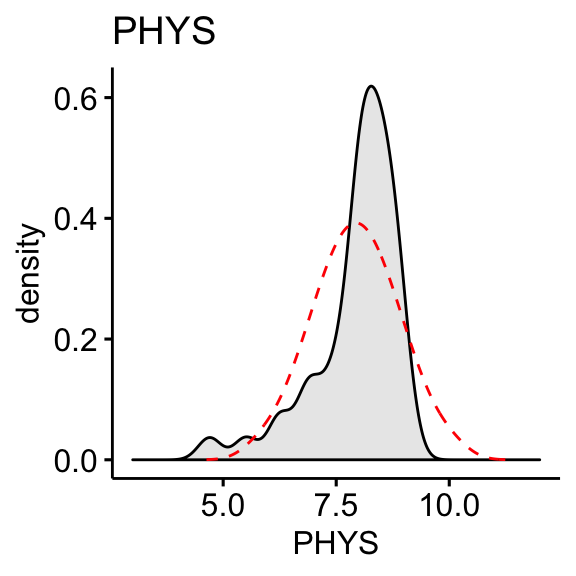
The “CONT” variable shows positive skewness. “PHYS” variable is negatively skewed
Compute skewness:
skewness(df$CONT, na.rm = TRUE)## [1] 1.09skewness(df$PHYS, na.rm = TRUE)## [1] -1.56Log transformation of the skewed data:
df$CONT <- log10(df$CONT)
df$PHYS <- log10(max(df$CONT+1) - df$CONT)# Distribution of CONT variable
ggdensity(df, x = "CONT", fill = "lightgray", title = "CONT") +
stat_overlay_normal_density(color = "red", linetype = "dashed")
# Distribution of PHYS variable
ggdensity(df, x = "PHYS", fill = "lightgray", title = "PHYS") +
stat_overlay_normal_density(color = "red", linetype = "dashed")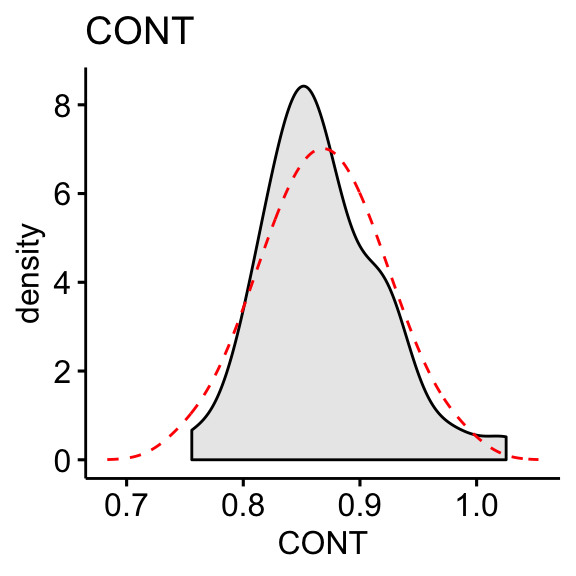
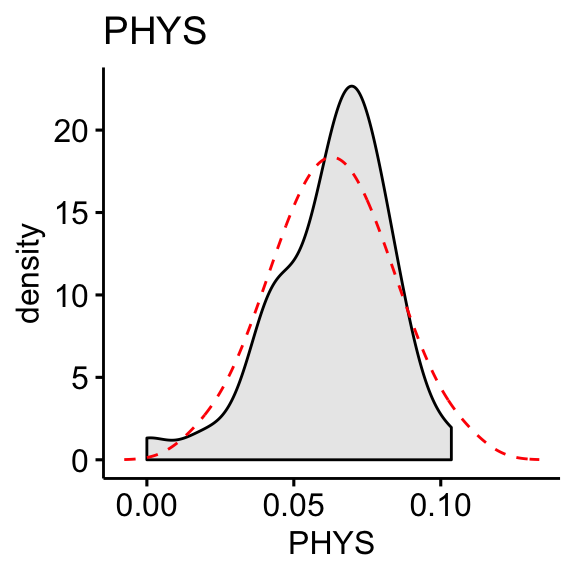
Compute skewness on the transformed data:
skewness(df$CONT, na.rm = TRUE)## [1] 0.656skewness(df$PHYS, na.rm = TRUE)## [1] -0.818The log10 transformation improves the distribution of the data to normality.
Summary and discussion
This article describes how to transform data for normality, an assumption required for parametric tests such as t-tests and ANOVA tests.
In the situation where the normality assumption is not met, you could consider running the statistical tests (t-test or ANOVA) on the transformed and non-transformed data to see if there are any meaningful differences.
If both tests lead you to the same conclusions, you might not choose to transform the outcome variable and carry on with the test outputs on the original data.
Note that transformation makes the interpretation of the analysis much more difficult. For example, if you run a t-test for comparing the mean of two groups after transforming the data, you cannot simply say that there is a difference in the two groups’ means. Now, you have the added step of interpreting the fact that the difference is based on the log transformation. For this reason, transformations are usually avoided unless necessary for the analysis to be valid.
For analyses like the F or t family of tests (i.e., independent and dependent sample t-tests, ANOVAs, MANOVAs, and regressions), violations of normality are not usually a death sentence for validity. With large enough sample sizes (> 30 or 40), there’s a pretty good chance that the data will be normally distributed; or at least close enough to normal that you can get away with using parametric tests (central limit theorem).
Recommended for you
This section contains best data science and self-development resources to help you on your path.
Books - Data Science
Our Books
- Practical Guide to Cluster Analysis in R by A. Kassambara (Datanovia)
- Practical Guide To Principal Component Methods in R by A. Kassambara (Datanovia)
- Machine Learning Essentials: Practical Guide in R by A. Kassambara (Datanovia)
- R Graphics Essentials for Great Data Visualization by A. Kassambara (Datanovia)
- GGPlot2 Essentials for Great Data Visualization in R by A. Kassambara (Datanovia)
- Network Analysis and Visualization in R by A. Kassambara (Datanovia)
- Practical Statistics in R for Comparing Groups: Numerical Variables by A. Kassambara (Datanovia)
- Inter-Rater Reliability Essentials: Practical Guide in R by A. Kassambara (Datanovia)
Others
- R for Data Science: Import, Tidy, Transform, Visualize, and Model Data by Hadley Wickham & Garrett Grolemund
- Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems by Aurelien Géron
- Practical Statistics for Data Scientists: 50 Essential Concepts by Peter Bruce & Andrew Bruce
- Hands-On Programming with R: Write Your Own Functions And Simulations by Garrett Grolemund & Hadley Wickham
- An Introduction to Statistical Learning: with Applications in R by Gareth James et al.
- Deep Learning with R by François Chollet & J.J. Allaire
- Deep Learning with Python by François Chollet
Version:
 Français
Français



Hi Kassambara,
another EXCELLENT, clear article.
But getting an error message from example:
# Distribution of CONT variable
ggdensity(df, x = “CONT”, fill = “lightgray”, title = “CONT”) +
scale_x_continuous(limits = c(3, 12)) +
stat_overlay_normal_density(color = “red”, linetype = “dashed”)
#returns error:
Error in data[, x] : object of type ‘closure’ is not subsettable
help!
oops! (Mea Culpa)
I missed a step in your Tutorial.
Ok, continue with the wonderful, clear examples…
OK, great that it works!
one more (typo) maybe?
at end of article, where it says:
df$PHYS <- log10(max(df$CONT+1) – df$CONT)
shouldn't say, instead ?:
df$PHYS <- log10(max(df$PHYS+1) – df$PHYS)
Then, the final:
skewness(df$PHYS, na.rm = TRUE)
will also have a different value.
Just asking…
Hello!
Would you have an academic reference (book, journal article) referring to a transformation in the form of log10(max(x+1) – x)?
Any hint would be most appreciated. Thank you!
Sincerely,
Mario
Hello Kassambara,
Thank you for this super helpful article,
At the end you mentioned that for ANOVA tests, violations of normality will not necessarily compromise validity; I was wondering if you were able to provide a reference for this? As it would help support my justification for not having to transform my non-normal data when I’m planning to perform an ANOVA.
Thank you!!